Books
Projects
Citation
Editorships
Molecular Simulations
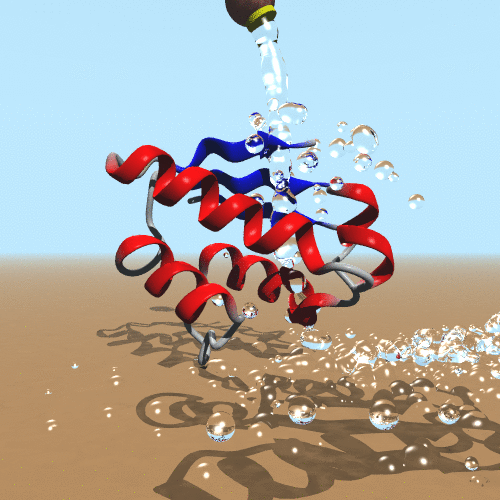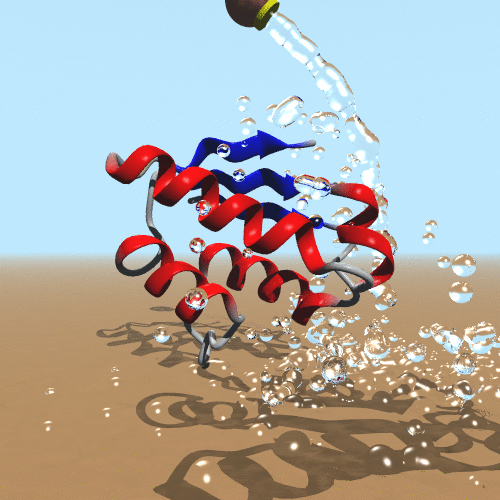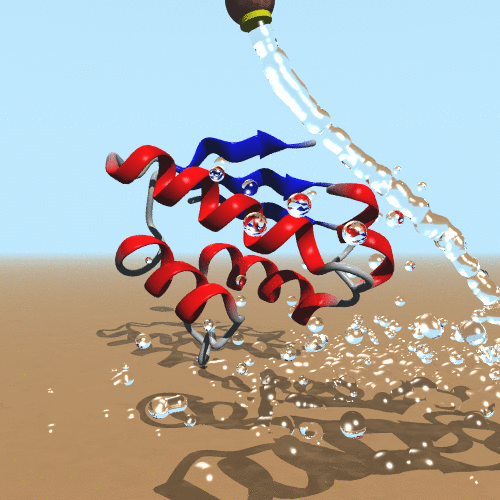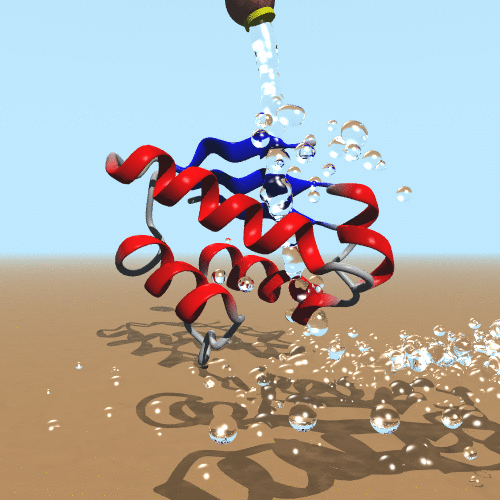 Our molecular world challenges us on an everyday basis to establish the theoretical foundations to understand it. From biological molecules to the next generation of nanomaterials, the possibility of simulating them is compelling. The impact of our scientific and technological development inspires a new generation of scientists to create new computational models to simulate molecules. The success of quantum mechanics makes it clear that it is our ultimate goal to simulate molecules. On the other hand, it is evident that computer power only limits simulations of polymers. Therefore, the development and application of classical and hybrid methodologies still have a beneficial impact to assess the behavior of molecules (de Azevedo & Dias, 2008). In our research, we focus on the development of a new generation of force fields targeted to the molecular system of interest (de Azevedo, 2011). We have been working on the development of new computational tools to study biomolecules (de Azevedo et al., 2001) and the interaction between them (Bitencourt-Ferreira & de Azevedo, 2019).
Our molecular world challenges us on an everyday basis to establish the theoretical foundations to understand it. From biological molecules to the next generation of nanomaterials, the possibility of simulating them is compelling. The impact of our scientific and technological development inspires a new generation of scientists to create new computational models to simulate molecules. The success of quantum mechanics makes it clear that it is our ultimate goal to simulate molecules. On the other hand, it is evident that computer power only limits simulations of polymers. Therefore, the development and application of classical and hybrid methodologies still have a beneficial impact to assess the behavior of molecules (de Azevedo & Dias, 2008). In our research, we focus on the development of a new generation of force fields targeted to the molecular system of interest (de Azevedo, 2011). We have been working on the development of new computational tools to study biomolecules (de Azevedo et al., 2001) and the interaction between them (Bitencourt-Ferreira & de Azevedo, 2019).
References
 Bitencourt-Ferreira G, de Azevedo WF Jr. Molecular Dynamics Simulations with NAMD2. Methods Mol Biol. 2019; 2053: 109–124. PubMed
Bitencourt-Ferreira G, de Azevedo WF Jr. Molecular Dynamics Simulations with NAMD2. Methods Mol Biol. 2019; 2053: 109–124. PubMed
de Azevedo WF Jr, Canduri F, Fadel V, Teodoro LG, Hial V, Gomes RA. Molecular model for the binary complex of uropepsin and pepstatin. Biochem Biophys Res Commun. 2001; 287(1): 277–281. PubMed
de Azevedo WF Jr, Dias R. Computational methods for calculation of ligand-binding affinity. Curr Drug Targets. 2008; 9(12): 1031–1039. PubMed
de Azevedo WF Jr. Molecular dynamics simulations of protein targets identified in Mycobacterium tuberculosis. Curr Med Chem. 2011; 18(9): 1353–1366. PubMed



 SAnDReS: Statistical Analysis of Docking Results and Scoring functions
SAnDReS: Statistical Analysis of Docking Results and Scoring functions


 Taba: A Tool to Analyze the Binding Affinity
Taba: A Tool to Analyze the Binding Affinity