Books
Projects
Citation
Editorships
SAnDReS
Highlights
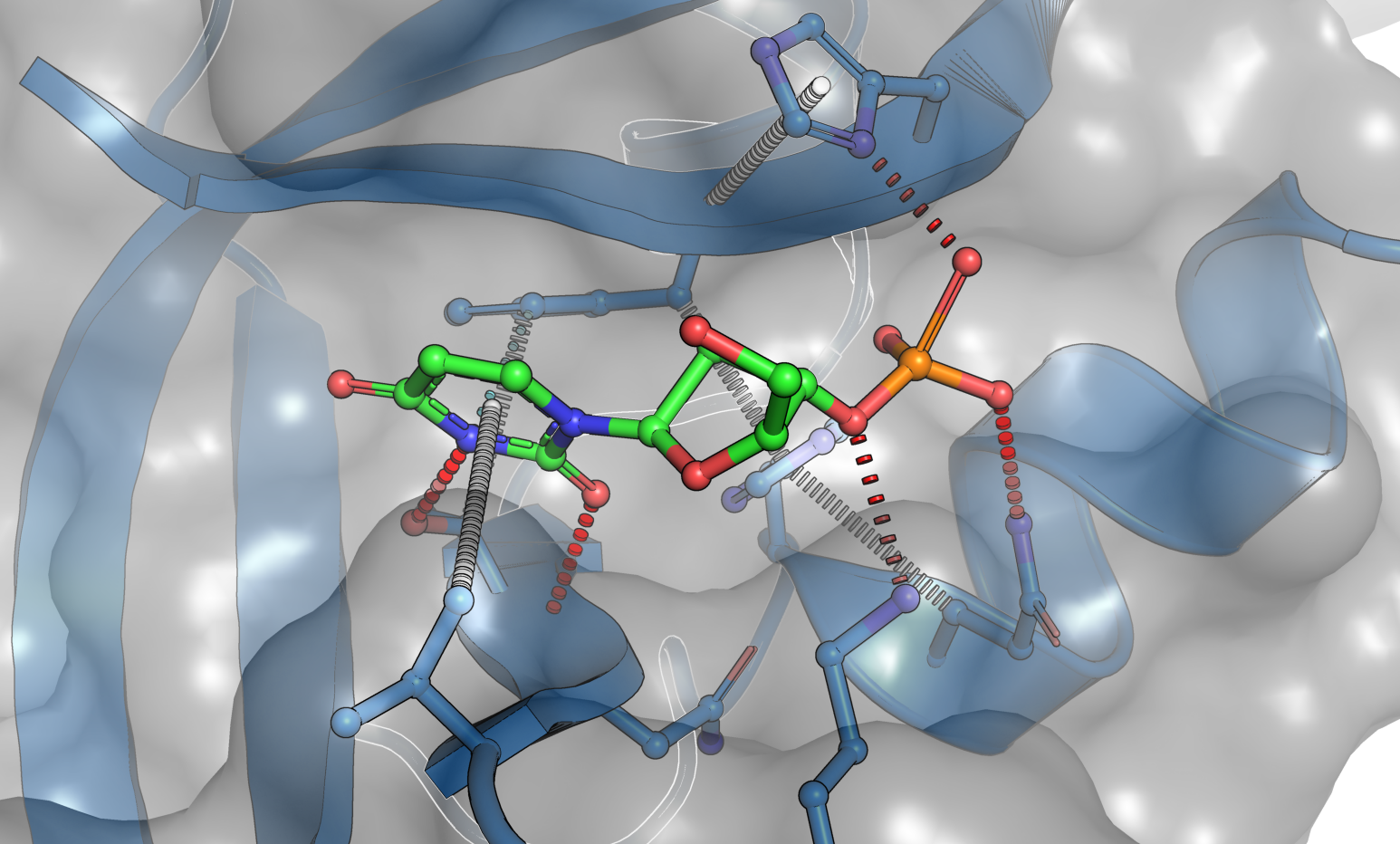
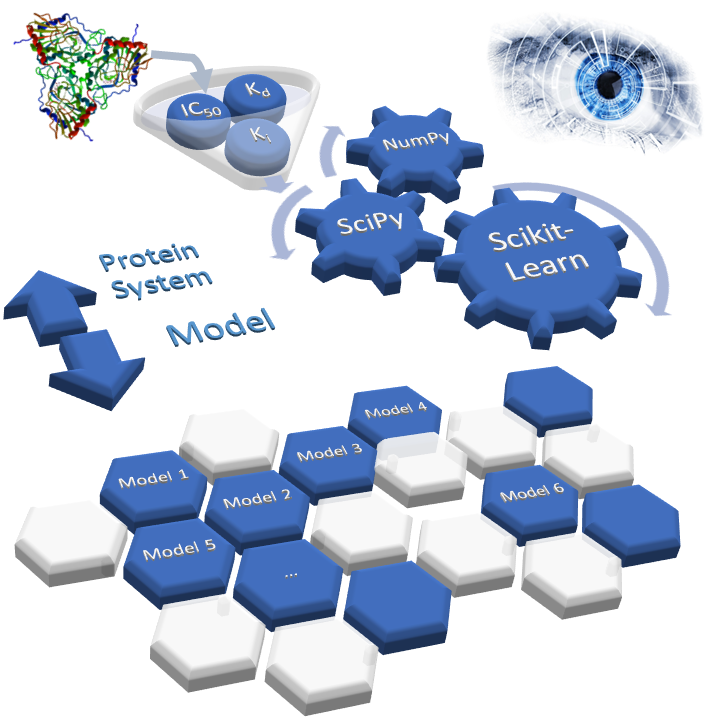 SAnDReS 2.0 (Statistical Analysis of Docking Results and Scoring functions) brings advanced computational tools for protein-ligand docking simulation and machine-learning modeling. We have AutoDock Vina (version 1.2.3) (Eberhardt et al., 2021) as a docking engine. Also, SAnDReS 2.0 has 54 regression methods implemented using Scikit-Learn (Pedregosa et al., 2011), which allows us to explore the Scoring Function Space (SFS) concept. This exploration of the SFS permits us to have an adequate machine-learning model for a targeted protein system. This approach creates computational models with superior predictive performance compared with classical scoring functions (also known as universal scoring functions). SAnDReS aims to merge the holistic view of systems biology with machine-learning methods to contribute to drug discovery projects. SAnDReS predicts binding affinity for a specific protein system with superior performance compared to classical scoring functions. Evaluation of the predictive performance of 107 scoring functions against the CASF-2016 benchmark (Su et al., 2019) indicates that a machine-learning model developed with SAnDReS 2.0 outperformed classical and machine-learning scoring functions such as KDEEP (Jiménez et al., 2018), CSM-lig (Pires & Ascher, 2016), and ΔVinaRF20 (Wang & Zhang, 2017) (plots below). Dr. Walter F. de Azevedo Jr. proposed the initial idea of SAnDReS in 2016, which now has an international team of scientists participating in its development and testing.
SAnDReS 2.0 (Statistical Analysis of Docking Results and Scoring functions) brings advanced computational tools for protein-ligand docking simulation and machine-learning modeling. We have AutoDock Vina (version 1.2.3) (Eberhardt et al., 2021) as a docking engine. Also, SAnDReS 2.0 has 54 regression methods implemented using Scikit-Learn (Pedregosa et al., 2011), which allows us to explore the Scoring Function Space (SFS) concept. This exploration of the SFS permits us to have an adequate machine-learning model for a targeted protein system. This approach creates computational models with superior predictive performance compared with classical scoring functions (also known as universal scoring functions). SAnDReS aims to merge the holistic view of systems biology with machine-learning methods to contribute to drug discovery projects. SAnDReS predicts binding affinity for a specific protein system with superior performance compared to classical scoring functions. Evaluation of the predictive performance of 107 scoring functions against the CASF-2016 benchmark (Su et al., 2019) indicates that a machine-learning model developed with SAnDReS 2.0 outperformed classical and machine-learning scoring functions such as KDEEP (Jiménez et al., 2018), CSM-lig (Pires & Ascher, 2016), and ΔVinaRF20 (Wang & Zhang, 2017) (plots below). Dr. Walter F. de Azevedo Jr. proposed the initial idea of SAnDReS in 2016, which now has an international team of scientists participating in its development and testing.
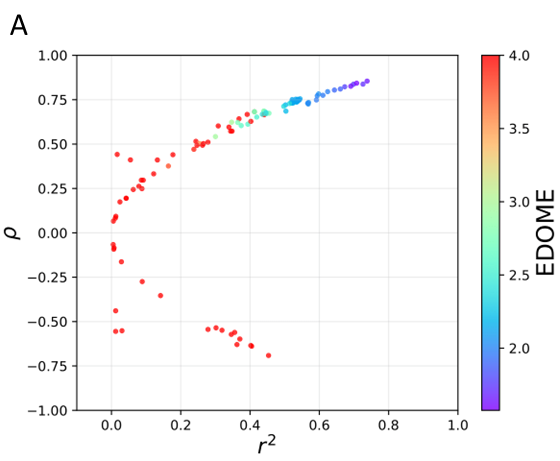
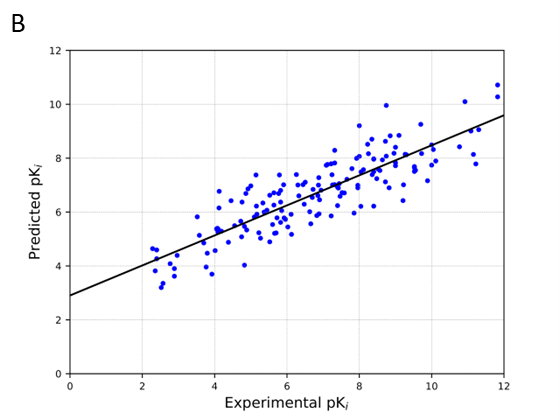
A) Predictive performance using DOME (Walsh et al., 2021) strategy (r2, rho, and EDOME). B) Scattering plot (Predicted pKi vs. Experimental pKi) for all docked structures in the CASF-2016 Ki test set.
Funding
The Brazilian National Council for Scientific and Technological Development (CNPq) (Process 306298/2022-8) supports this research project. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) – Finance Code 001. MVA acknowledges Diether Haenicke Scholarship from Western Michigan University. ОТ, NB, and VP thank the Program for Basic Research in the Russian Federation for a long-term period 2021–2030 (project No. 122030100170-5). R.Q and M.A.V thank Secyt-UNC for theirfinancial support.



 SAnDReS: Statistical Analysis of Docking Results and Scoring functions
SAnDReS: Statistical Analysis of Docking Results and Scoring functions


 Taba: A Tool to Analyze the Binding Affinity
Taba: A Tool to Analyze the Binding Affinity