Books
Projects
Citation
Editorships
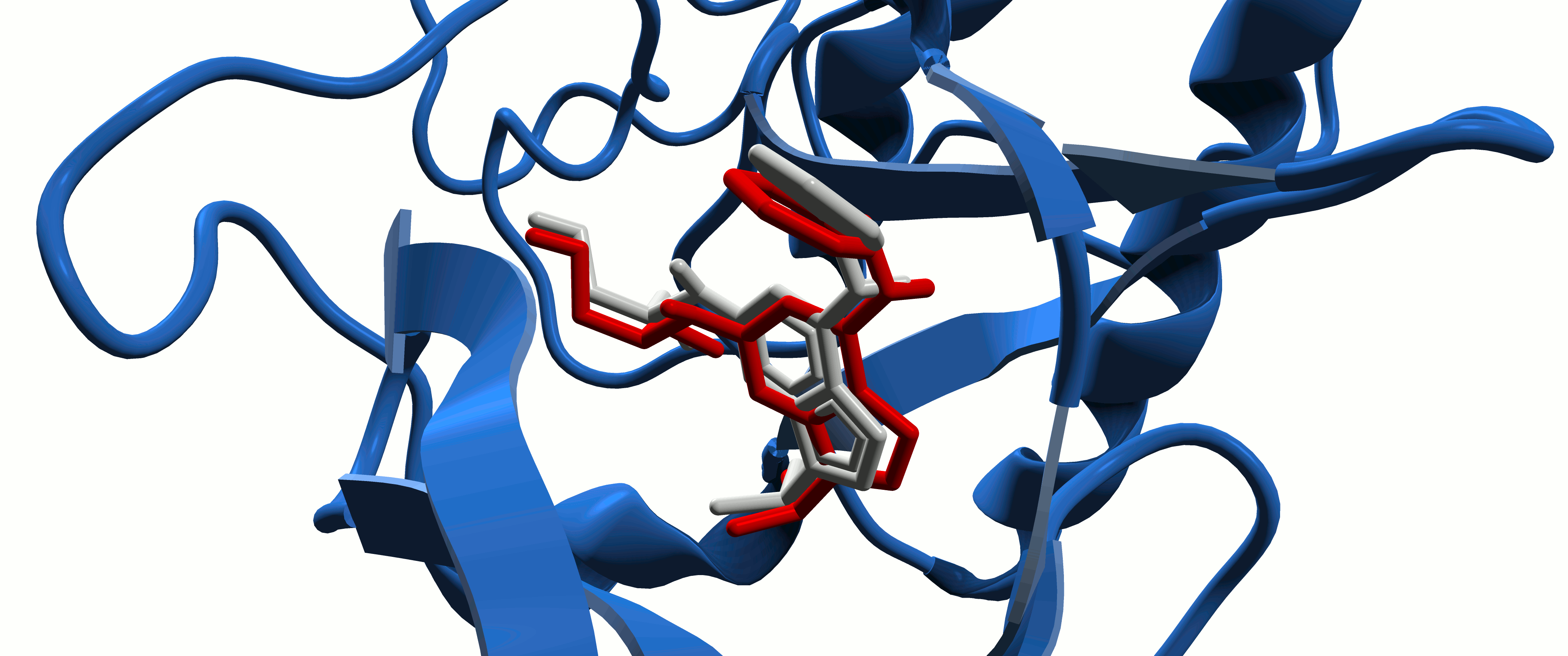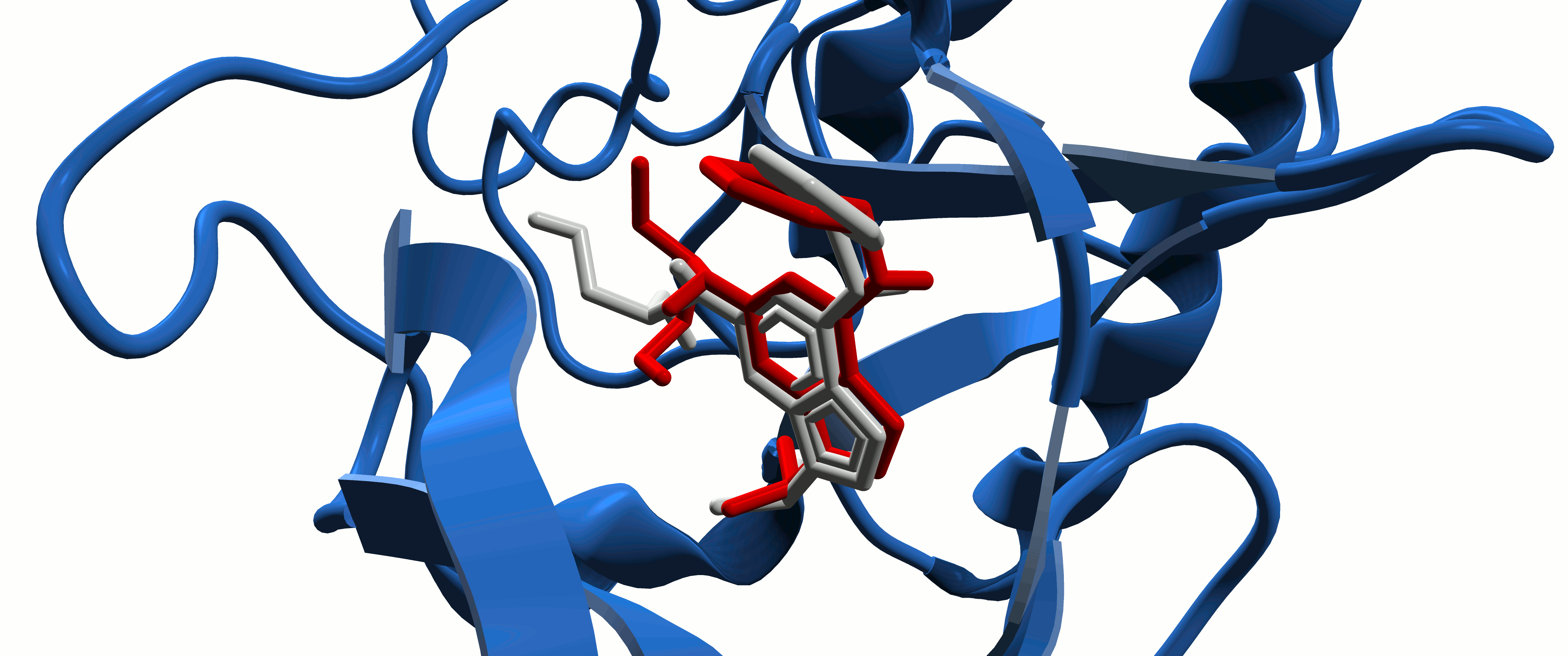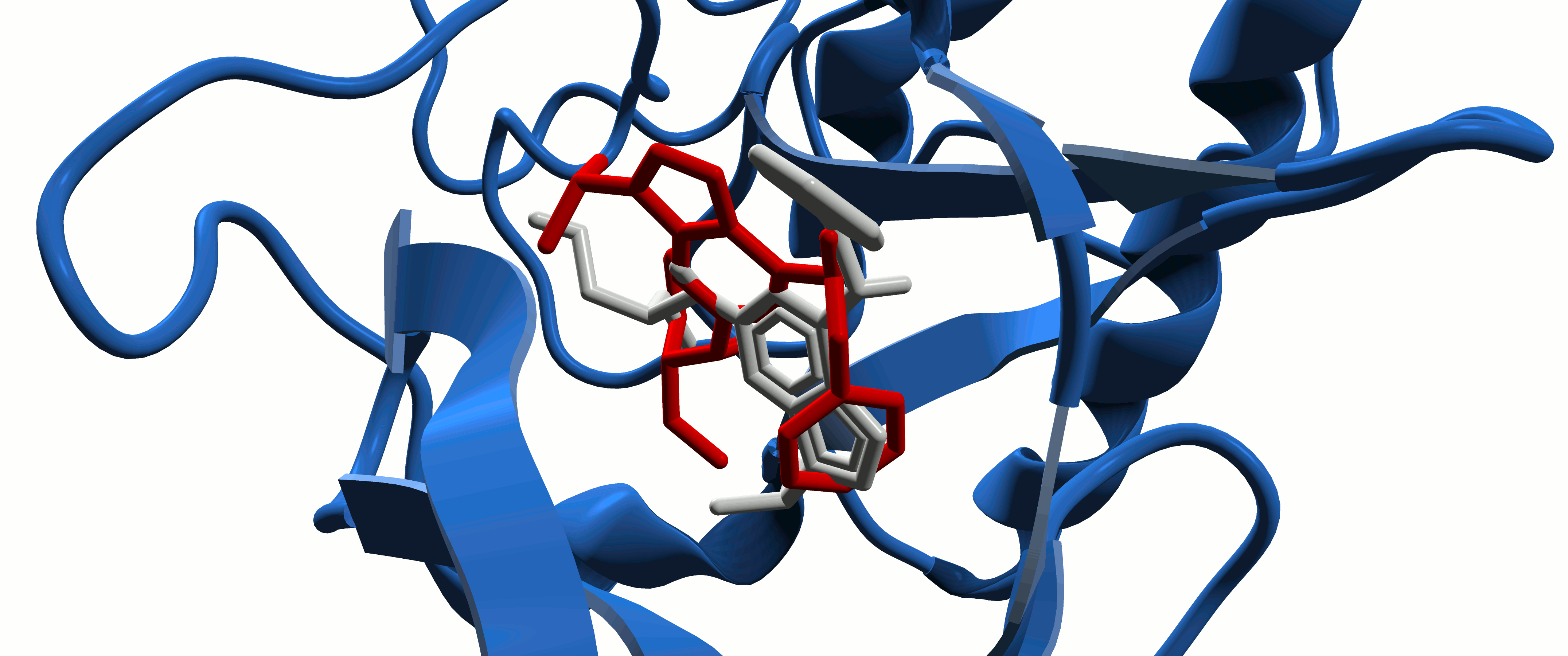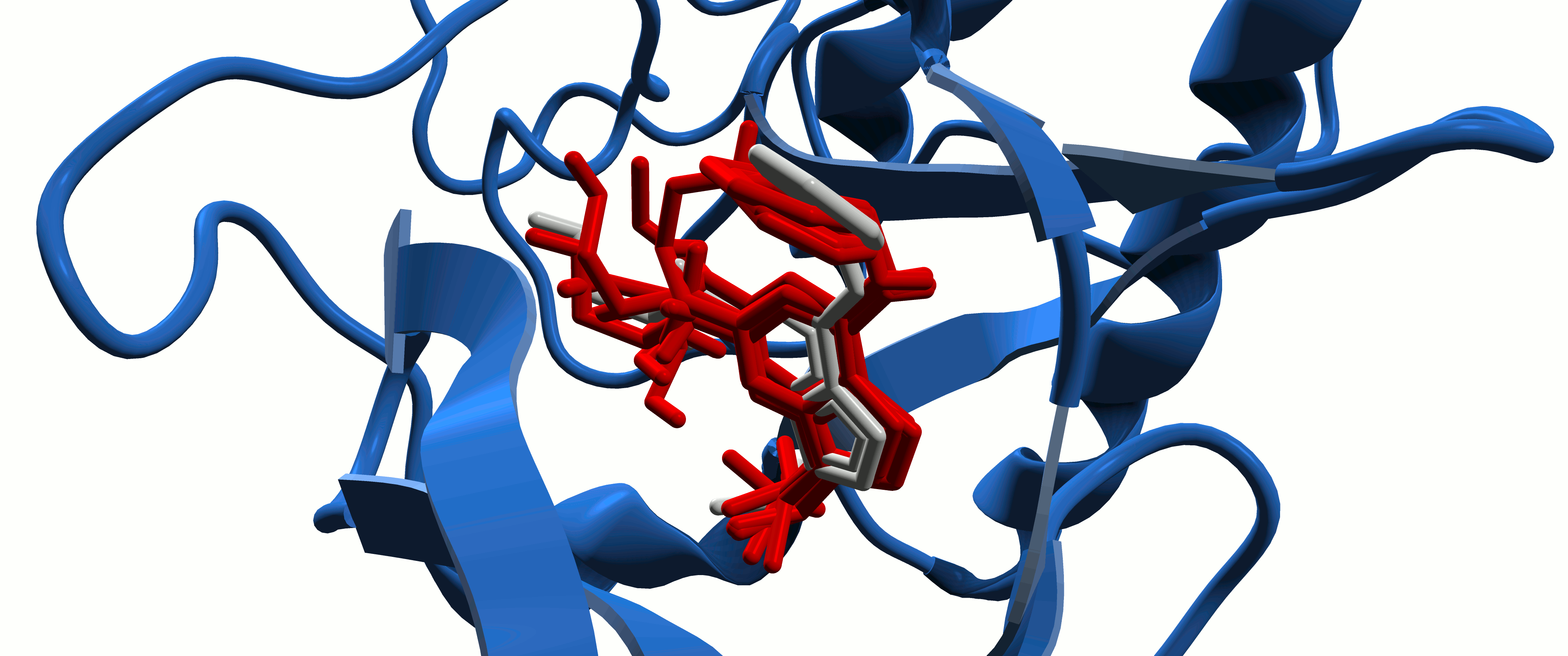
Protein-Ligand Interactions
In the study of intermolecular interactions involving protein and ligands, we expect to gain further insights into the structural basis for the specificity of small-molecule ligands against a specific protein target (de Azevedo, 2008). Analysis of protein-ligand interaction is a central problem in drug design. Knowledge of the key features responsible for the specificity of a ligand for a protein allows us to determine which physical-chemical parameters could improve the protein-ligand interaction (de Azevedo & Dias, 2008a). Furthermore, the development of a computational model to predict the binding affinity based on the atomic coordinates of a protein-ligand complex opens the possibility to apply virtual screening approaches to search small-molecule databases to identify a drug candidate (de Azevedo & Dias, 2008b, de Azevedo, 2010a; Bitencourt-Ferreira & de Azevedo, 2019a; da Silva et al., 2020 ). To study protein-ligand interactions, we make use of protein crystallography (Canduri & de Azevedo, 2008; Veit-Acosta & de Azevedo, 2021), nuclear magnetic resonance spectroscopy (Fadel et al., 2005), molecular docking (de Azevedo, 2010b), and molecular dynamics (de Azevedo, 2011; Bitencourt-Ferreira & de Azevedo, 2019b).
References
Canduri F, de Azevedo WF. Protein crystallography in drug discovery. Curr Drug Targets. 2008; 9(12):1048–1053. PubMed
de Azevedo WF Jr. Protein-drug interactions. Curr Drug Targets. 2008; 9(12):1030. PubMed
de Azevedo WF Jr, Dias R. Experimental approaches to evaluate the thermodynamics of protein-drug interactions. Curr Drug Targets. 2008a; 9(12):1071–1076. PubMed
de Azevedo WF Jr, Dias R. Computational methods for calculation of ligand-binding affinity. Curr Drug Targets. 2008b; 9(12):1031–1039. PubMed
de Azevedo WF Jr. Structure-based virtual screening. Curr Drug Targets. 2010a; 11(3):261–263. PubMed
de Azevedo WF Jr. MolDock applied to structure-based virtual screening. Curr Drug Targets. 2010b; 11(3):327–334. PubMed
de Azevedo WF Jr. Molecular dynamics simulations of protein targets identified in Mycobacterium tuberculosis. Curr Med Chem. 2011; 18(9):1353–1366. PubMed
Fadel V, Bettendorff P, Herrmann T, de Azevedo WF Jr, Oliveira EB, Yamane T, Wüthrich K. Automated NMR structure determination and disulfide bond identification of the myotoxin crotamine from Crotalus durissus terrificus. Toxicon. 2005; 46(7):759–767. PubMed
Veit-Acosta M, de Azevedo Junior WF. The Impact of Crystallographic Data for the Development of Machine Learning Models to Predict Protein-Ligand Binding Affinity. Curr Med Chem. doi: 10.2174/0929867328666210210121320. PubMed



 SAnDReS: Statistical Analysis of Docking Results and Scoring functions
SAnDReS: Statistical Analysis of Docking Results and Scoring functions


 Taba: A Tool to Analyze the Binding Affinity
Taba: A Tool to Analyze the Binding Affinity







 d
d