Books
Projects
Citation
Editorships
Taba
Highlights
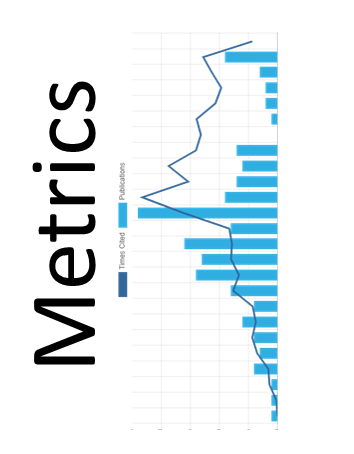
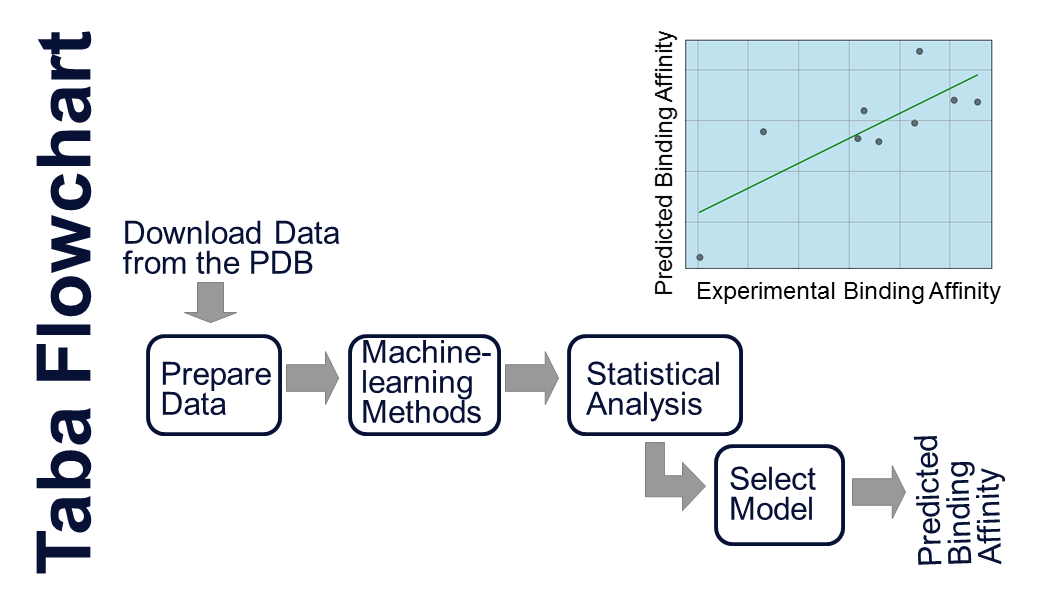
Taba (Tool to Analyze the Binding Affinity) generates scoring functions to predict binding affinity based on the atomic coordinates of a protein-ligand complex. It employs a polynomial equation where the terms are mass-spring potentials. Taba calculates average interatomic distances and takes them as equilibrium distances for a mass-spring potential. The mass-spring potential equation and the equilibrium constants for each pair of atoms compose the Taba Force Field (TFF). We are currently updating Taba and expect to release a new version of Taba by July/2024. It is indicated below a flowchart highlighting the main steps of Taba. Please cite the following reference (da Silva et al., 2020) if you use the Taba program.
This flowchart shows the main steps used to generate targeted-scoring functions with Taba (da Silva et al., 2020).
TFF outperforms classical and machine learning scoring functions such as KDEEP (Jiménez et al., 2018) and CSM-lig (Pires & Ascher, 2016)(plot below).
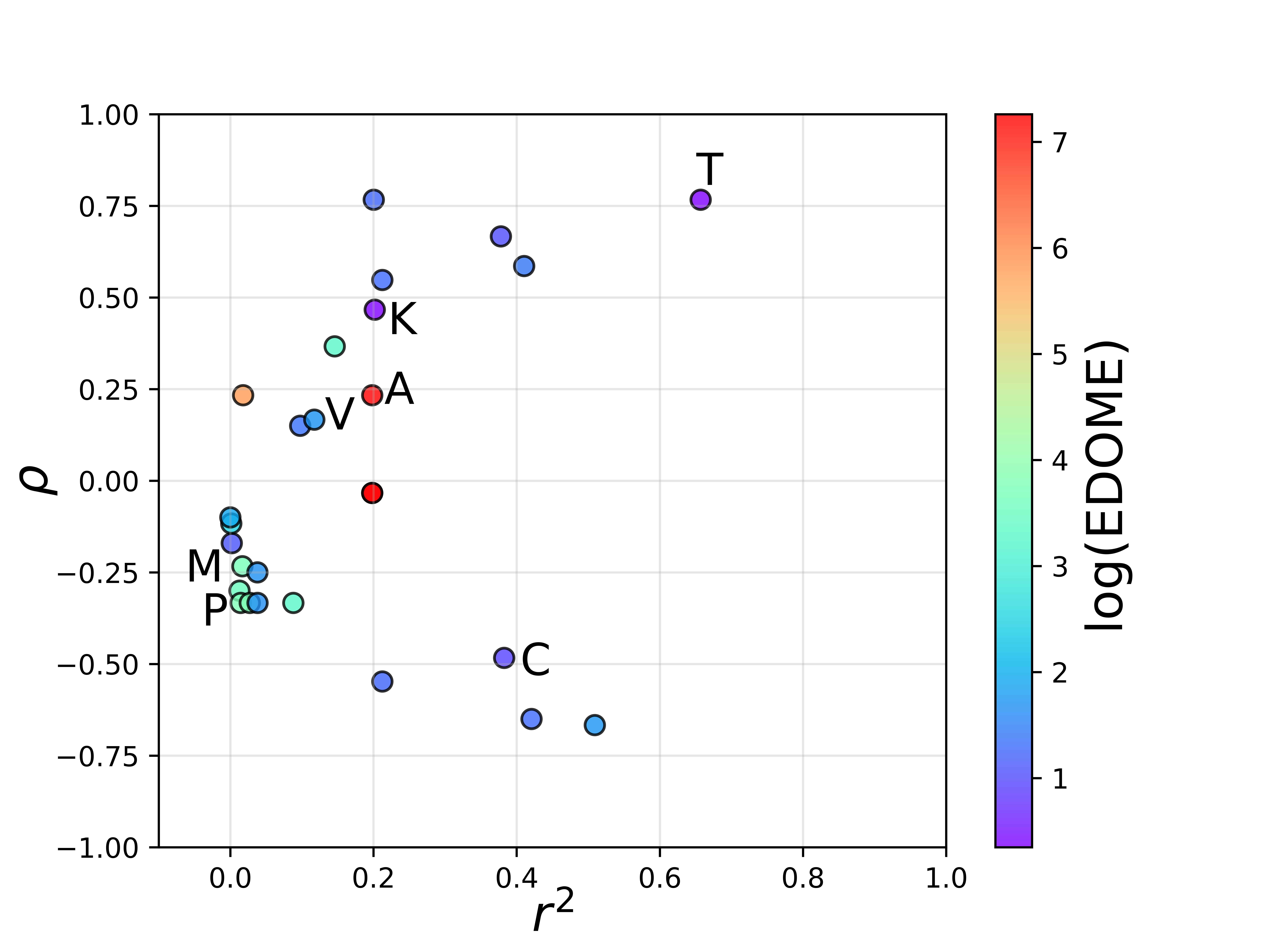
Predictive performance with DOME (Walsh et al., 2021) using r2, rho, and log (EDOME). We employed EDOME to generate this plot. Statistical analysis of scoring functions includes previously published data (da Silva et al., 2020) to create this plot, except for KDEEP (Jiménez et al., 2018) and CSM-lig (Pires & Ascher, 2016), which utilized the atomic coordinates for the structures in the test set to predict the affinity (pKi). The plot highlights the performance of seven scoring functions: T (Taba), K (KDEEP ), C (CSM-lig), A (AutoDock4) (Morris et al. 2009), V (AutoDock Vina) (Eberhardt et al., 2021), M (MolDock Score), and P (Plants Score) (Thomsen & Christensen, 2006).
Downloads
Funding
The Brazilian National Council for Scientific and Technological Development (CNPq) (Process 306298/2022-8) supports this research project. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) – Finance Code 001. MVA acknowledges Diether Haenicke Scholarship from Western Michigan University. ОТ, NB, and VP thank the Program for Basic Research in the Russian Federation for a long-term period 2021–2030 (project No. 122030100170-5). R.Q and M.A.V thank Secyt-UNC for their financial support.



 SAnDReS: Statistical Analysis of Docking Results and Scoring functions
SAnDReS: Statistical Analysis of Docking Results and Scoring functions


 Taba: A Tool to Analyze the Binding Affinity
Taba: A Tool to Analyze the Binding Affinity