Books
Projects
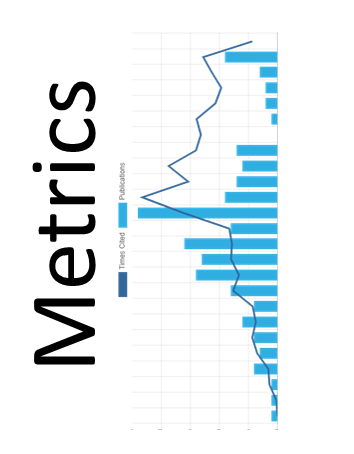
Citation
Editorships
Dr. Walter F. de Azevedo, Jr. has been ranked among the most influential researchers in the world according to a database created by Journal Plos Biology (see news here). Dr. Azevedo`s influential works can be found here. Citation metrics available here.
Updates
- August 2021 data-update for "Updated science-wide author databases of standardized citation indicators"
- September 2022 data-update for "Updated science-wide author databases of standardized citation indicators"
- October 2023 data-update for "Updated science-wide author databases of standardized citation indicators"
Aula Magna [>>]
Exploring the Scoring Function Space [>>]
A view of the scoring function space as a way to develop a computational model to predict ligand-binding affinity.
We envisage protein-ligand interaction as a result of the relation between the protein space (Smith, 1970; Hou et al., 2005) and the chemical space (Bohacek et al., 1996; Dobson, 2004; Kirkpatrick & Ellis, 2004; Lipinski & Hopkins, 2004; Shoichet, 2004; Stockwell, 2004), and we propose to approach these sets as a complex system, where the application of computational methodologies could contribute to understanding the structural basis for the specificity of ligands for proteins. Such approaches can create novel scoring functions to predict binding affinity with superior predictive power compared with classical scoring functions (also known as universal scoring functions)(Ross et al., 2013) available in docking programs (de Azevedo Jr., 2021). We propose to use the abstraction of a mathematical space composed of infinite computational models to predict ligand-binding affinity, named scoring function space (SFS) (Ross et al., 2013; Heck et al., 2017; Bitencourt-Ferreira & de Azevedo Jr., 2019; Veríssimo et al., 2022). With the development of the SFS concept, we expect to merge the holistic view of systems biology with machine-learning methods to contribute to drug discovery projects (Bitencourt-Ferreira et al., 2023). By the use of supervised machine learning techniques, we can explore this SFS (Bitencourt-Ferreira & de Azevedo Jr., 2019; Bitencourt-Ferreira et al., 2023) to build a computational model targeted to a specific protein system (targeted-scoring function) (Seifert, 2009). For instance, we created targeted-scoring functions for cyclin-dependent kinases (EC 2.7.11.22) (de Ávila et al., 2017; Levin et al., 2018) and HIV-1 protease (EC 3.4.23.16) (Pintro & de Azevedo, 2017). We developed the programs SAnDReS (Xavier et al., 2016; Bitencourt-Ferreira & de Azevedo Jr., 2019), SFSXplorer, and Taba (da Silva et al., 2020; Bitencourt-Ferreira et al., 2021) to generate computational models to predict ligand-binding affinity. These programs are integrated computational tools to explore the SFS.
Impact [>>]
- Bio
- Books
- Citation Metrics
- Editorships
- Most Influential Researchers in the World
- News
- Publications
- Talks
"Let the light of science end the darkness of denialism."
If you have interest in participating in our research projects, please send an e-mail in English to research@azevedolab.net



 SAnDReS: Statistical Analysis of Docking Results and Scoring functions
SAnDReS: Statistical Analysis of Docking Results and Scoring functions


 Taba: A Tool to Analyze the Binding Affinity
Taba: A Tool to Analyze the Binding Affinity