Books
Projects
Citation
Editorships
Computational Systems Biology
As highlighted in a seminal paper by Kitano, 2002: "Computational systems biology addresses questions fundamental to our understanding of life, yet progress here will lead to practical innovations in medicine, drug discovery, and engineering." With this view, we aim to merge the holistic approach of systems biology with machine-learning methods to contribute to drug discovery projects (Bitencourt-Ferreira et al., 2023). Voit says biological systems generally contain several units, but they present an additional challenge to any study because the processes that govern them are not linear (Voit, 2018). For instance, the use of Biochemical Systems Theory (Voit & Savageau, 1987) allows us to model complex biological systems to predict oscillatory patterns (e.g., cyclin-dependent kinase 2) (Goldbeter, 2013) (Figure below) and other behaviors.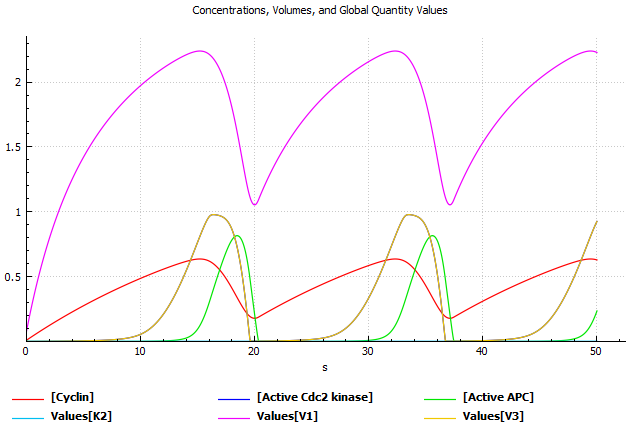
Model for oscillations of Cdc kinase activity in embryonic cell cycles based on Michaelis-Menten phosphorylation-dephosphorylation kinetics (Goldbeter, 2013) (SBML model available here). Square brackets indicate concentrations. Vi denotes the effective maximum rate for each of the enzymes. APC means anaphase-promoting complex. COPASI (Mendes et al., 2009) generated the above plot.
We have been working on computational models to predict enzyme inhibition and protein-ligand interactions (Bitencourt-Ferreira & de Azevedo, 2019a; 2019b; 2019c; da Silva et al., 2020). We can use these computational models to predict the binding affinity of a potential inhibitor for an enzyme; such knowledge speeds up drug discovery and decreases the cost of new drugs (de Ávila et al., 2017; Pintro & Azevedo, 2017).
Furthermore, the availability of computational models to predict binding affinity based on the atomic coordinates of protein-ligand complexes adds flexibility to drug discovery (Xavier et al., 2016; Heck et al., 2017). It allows us to computationally test different scenarios where a potential new drug may interact with a protein target (Bitencourt-Ferreira & de Azevedo, 2021). We developed the programs SAnDReS (Xavier et al., 2016; Bitencourt-Ferreira et al., 2021) and Taba (da Silva et al., 2020, Bitencourt-Ferreira et al., 2021) to create machine-learning models targeted to the protein system of interest. We have successfully employed SAnDReS to study coagulation factor Xa (EC 3.4.21.6) (Xavier et al., 2016), cyclin-dependent kinases (EC 2.7.11.22) (de Ávila et al., 2017; Levin et al., 2018), HIV-1 protease (EC 3.4.23.16) (Pintro & de Azevedo, 2017), estrogen receptor (Amaral et al., 2018), cannabinoid receptor 1 (Russo & de Azevedo, 2019; Russo & de Azevedo, 2020), and 3-dehydroquinate dehydratase (EC 4.2.1.10) (de Ávila & de Azevedo, 2018). Also, we used SAnDReS (Xavier et al., 2016) to develop a machine-learning model to predict Gibbs free energy of binding for protein-ligand complexes (Bitencourt-Ferreira & de Azevedo Jr., 2018).
References
Amaral MEA, Nery LR, Leite CE, de Azevedo Junior WF, Campos MM. Pre-clinical effects of metformin and aspirin on the cell lines of different breast cancer subtypes. Invest New Drugs. 2018; 36(5): 782–796. PubMed
Bitencourt-Ferreira G, de Azevedo Jr. WF. Development of a machine-learning model to predict Gibbs free energy of binding for protein-ligand complexes. Biophys Chem. 2018; 240: 63–69. PubMed
Bitencourt-Ferreira G, de Azevedo WF Jr. Machine Learning to Predict Binding Affinity. Methods Mol Biol. 2019a; 2053: 251–273. PubMed
Bitencourt-Ferreira G, de Azevedo WF Jr. Exploring the Scoring Function Space. Methods Mol Biol. 2019b; 2053: 275–281. PubMed
Bitencourt-Ferreira G, de Azevedo WF Jr. How Docking Programs Work. Methods Mol Biol. 2019c; 2053: 35–50. PubMed
Bitencourt-Ferreira G, da Silva AD, de Azevedo WF Jr. Application of Machine Learning Techniques to Predict Binding Affinity for Drug Targets. A Study of Cyclin-Dependent Kinase 2. Curr Med Chem. 2021; 28(2): 253–265. PubMed
Bitencourt-Ferreira G, Rizzotto C, de Azevedo Junior WF. Machine Learning-Based Scoring Functions. Development and Applications With SAnDReS. Curr Med Chem. 2021; 28(9): 1746–1746. PubMed
Bitencourt-Ferreira G, de Azevedo Junior WF. Electrostatic Potential Energy in Protein-Drug Complexes. Curr Med Chem. 2021; 28(24): 4954–4971. PubMed
Bitencourt-Ferreira G, Villarreal MA, Quiroga R, Biziukova N, Poroikov V, Tarasova O, de Azevedo Junior WF. Exploring Scoring Function Space: Developing Computational Models for Drug Discovery. Curr Med Chem. 2023 Mar 21. doi: 10.2174/0929867330666230321103731. Epub ahead of print. PMID: 36944627. PubMed
Da Silva AD, Bitencourt-Ferreira G, de Azevedo WF Jr. Taba: A Tool to Analyze the Binding Affinity. J Comput Chem. 2020; 41(1): 69–73. PubMed
De Ávila MB, Xavier MM, Pintro VO, de Azevedo WF. Supervised machine learning techniques to predict binding affinity. A study for cyclin-dependent kinase 2. Biochem Biophys Res Commun. 2017; 494: 305–310. PubMed
De Ávila MB, de Azevedo WF Jr. Development of machine learning models to predict inhibition of 3-dehydroquinate dehydratase. Chem Biol Drug Des. 2018; 92: 1468–1474. PubMed
Goldbeter A. Oscillatory enzyme reactions and Michaelis-Menten kinetics. FEBS Lett. 2013; 587(17): 2778–2784. PubMed
Heck GS, Pintro VO, Pereira RR, de Ávila MB, Levin NMB, de Azevedo WF. Supervised Machine Learning Methods Applied to Predict Ligand-Binding Affinity. Curr Med Chem. 2017; 24(23): 2459–2470. PubMed
Kitano, H. Computational systems biology. Nature 2002; 420: 206–210. PubMed
Levin NMB, Pintro VO, Bitencourt-Ferreira G, Mattos BB, Silvério AC, de Azevedo Jr. WF. Development of CDK-targeted scoring functions for prediction of binding affinity. Biophys Chem. 2018; 235: 1–8. PubMed
Mendes P, Hoops S, Sahle S, Gauges R, Dada J, Kummer U. Computational modeling of biochemical networks using COPASI. Methods Mol Biol. 2009; 500: 17–59. PubMed
Pintro VO, Azevedo WF. Optimized Virtual Screening Workflow. Towards Target-Based Polynomial Scoring Functions for HIV-1 Protease. Comb Chem High Throughput Screen. 2017; 20(9): 820–827. PubMed
Russo S, De Azevedo WF. Advances in the Understanding of the Cannabinoid Receptor 1 - Focusing on the Inverse Agonists Interactions. Curr Med Chem. 2019; 26(10): 1908–1919. PubMed
Russo S, de Azevedo WF Jr. Computational Analysis of Dipyrone Metabolite 4-Aminoantipyrine as a Cannabinoid Receptor 1 Agonist. Curr Med Chem. 2020; 27(28): 4741–4749. PubMed
Voit EO, Savageau MA. Accuracy of alternative representations for integrated biochemical systems. Biochemistry. 1987; 26(21): 6869–6880. PubMed
Voit EO. A first course in systems biology. 2nd ed. New York (USA): Garland Science; c2018. Chapter 1, Biological systems; p. 1-18. DOI: https://doi.org/10.4324/9780203702260 Link
Voit, E. O. (2018). A first course in Systems Biology (2nd ed.). Garland Science.
Xavier MM, Heck GS, de Avila MB, Levin NM, Pintro VO, Carvalho NL, Azevedo WF Jr. SAnDReS a Computational Tool for Statistical Analysis of Docking Results and Development of Scoring Functions. Comb Chem High Throughput Screen. 2016; 19(10): 801–812. PubMed



 SAnDReS: Statistical Analysis of Docking Results and Scoring functions
SAnDReS: Statistical Analysis of Docking Results and Scoring functions


 Taba: A Tool to Analyze the Binding Affinity
Taba: A Tool to Analyze the Binding Affinity